Research Interests
I study mechanisms that generate, maintain, and preserve biodiversity.
- The ecology of species interactions
- Hierarchical Bayesian models for ecological predictions
- Computer vision and machine learning for biodiversity image analysis
Deep Learning for Biodiversity Detection
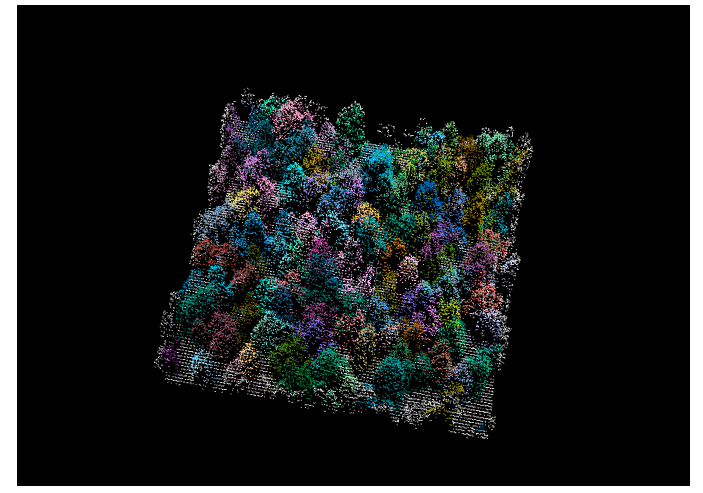
I recently joined the weecology lab at the University of Florida to work on pioneering research for deep learning for biodiversity detection in remotely sensed imagery. Our first project focuses on combining LIDAR, hyperspectral and rgb data for tree segmentation and classification.
Demo: http://tree.westus.cloudapp.azure.com/trees/
Demo: http://tree.westus.cloudapp.azure.com/trees/
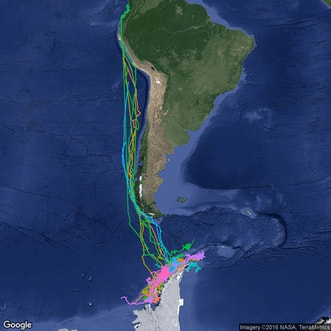
Animal movement in dynamic landscapes
Using new satellite tracking technology, we are studying optimal foraging and the ecological predictors of animal movement.
Check out my interactive tools to visualize humpback whale movement and migration.
Check out my interactive tools to visualize humpback whale movement and migration.
Specialization and Niche Overlap in Andean Hummingbirds
I study co-evolution and niche partitioning of tropical plants and hummingbirds in montane cloud forests.
Live Data Dashboard: https://weinstein.shinyapps.io/HummingbirdData/
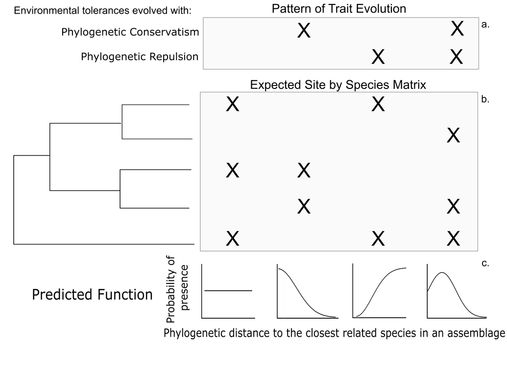
Co-occurrence and relatedness in ecological assemblages
|
Graham LJ, Weinstein BG, Supp SR, Graham CH. Future geographic patterns of novel and disappearing assemblages across three dimensions of diversity: A case study with Ecuadorian hummingbirds. Divers. Distrib. 2017;00:1–11. https://doi.org/10.1111/ddi.12587
Lessard, J.P., Weinstein, B.G., Borregaard, M.K., Marske, K.A., Martin, D.R., McGuire, J.A., Parra, J.L., Rahbek, C. and Graham, C.H., 2015. Process-Based Species Pools Reveal the Hidden Signature of Biotic Interactions Amid the Influence of Temperature Filtering. The American Naturalist, 187:75-88. Penone, C.*, Weinstein, B. G.*, Graham, C. H., Hedges, S. B., Rondonini, C., Davidson, A. and C. G. Costa. 2016. Global mammal betadiversity reveals convergence between isolated forest assemblages. Proceeding of the Royal Academy B: Biological Sciences. 283: 20161028. * authors contributed equally |
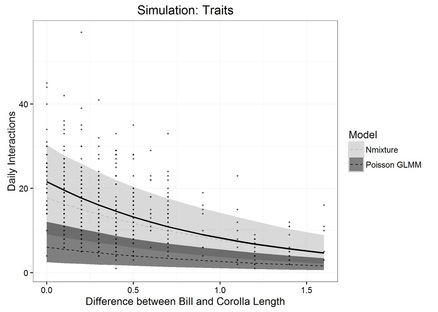
Hierarchical Bayesian Models for Species Interactions
Selectivity and Competitive Dominance
Harnessing Computer Vision for Ecological Monitoring
|
The cost, time, and logistics of human observation limit many studies, yet ecologists have only begun to use automated tools. Computer vision is a field of computer science that extracts information from images and combines data taken from the field with automated image analysis.
|
Weinstein, B.G. (2015) MotionMeerkat: integrating motion video detection and ecological monitoring Methods in Ecology and Evolution, 6, 357–362.
Cruzan, M.B, Weinstein, B. G., Grasty, M. R., Kohrn, B., Schroyer, T and Pamela G. Thompson. 2016. Small Unmanned Aerial Vehicles (Micro-UAVs -Drones) in Plant Ecology. Applications in Plant Sciences. 4: 1600041. |
Merging phylogeny and morphology with community ecology
Past Research
Predicting Latent Risk for Global Mammal Diversity
|
A collaboration with Dr. Ana Davidson and NatureServe studying the morphological correlates of risk in global mammal diversity. Combining phylogenetic and functional trait data with machine learning models to identify the life-history and spatial characteristics of mammalian biodiversity risk.
|
Davidson, A. D., Shoemaker, K. T., Weinstein, B. G., Costa, G. C., Brooks, T. M., Ceballos, G., Radeloff, V. C., Rondinini, C. and C. H. Graham. Geography of current and future global mammal extinction risk. PlosONE. Accepted.
|
Methods in Landscape Genetics and Phylobetadiversity

Using a mechanistic energy budget (Gifford and Kozak 2012) of Red-cheeked Salamanders (left), i'm using least cost path approaches to explain population genetic differentiation.This research is in conjunction with Amy Luxbacher at the University of Minnesota as part of her doctoral work.